
Manual
Download
Installation
FPB has been tested on ABI 3730 files. Fingerprints from different machines and file formats may be processed as long as similar input is provided, see below.
FPB takes in input text files output by GeneMapper only!
Such files can be produced via ABI GeneMapper manual export into tab-delimited text tables files or via GeneMapper batch export into tab-delimited text table files (see GeneMapper manual, including the Appendix). Alternatively you can produce similar files with any format and then convert them to GeneMapper tab-delimited text files writing a simple Perl script.
Note: when you export fragment sizes into files from GeneMapper, please do not make any editing and keep all fragments in the output files. FPB will handle the fragment editing.
GeneMapper software can create one output text file (*.dat) for all sample file (*.fsa) in one or more plates with the following or similar format (each line containing 6 columns separated by a tab):
Dye/Sample Peak Sample File Name Size Height Area Data Point "B,1" Lib5_Plate173_A01.fsa 13 49 10 "B,2" Lib5_Plate173_A01.fsa 11 41 79 "B,3" Lib5_Plate173_A01.fsa 10 40 124 "B,4" Lib5_Plate173_A01.fsa 12 83 135 ... "B,124" Lib5_Plate173_A01.fsa 76.61 219 1452 2145 "B,125" Lib5_Plate173_A01.fsa 79.4 247 813 2164 "B,126" Lib5_Plate173_A01.fsa 81.45 960 4012 2178 "B,127" Lib5_Plate173_A01.fsa 82.62 2918 12864 2186 ... "G,238" Lib5_Plate173_A01.fsa 203.52 380 1795 3052 "G,239" Lib5_Plate173_A01.fsa 204.82 695 6125 3062 "G,240" Lib5_Plate173_A01.fsa 206.26 627 3390 3073 "G,241" Lib5_Plate173_A01.fsa 207.04 264 1791 3079 "G,242" Lib5_Plate173_A01.fsa 208.86 275 1247 3093 ... "R,152" Lib5_Plate173_A01.fsa 94.16 266 1094 2265 "R,153" Lib5_Plate173_A01.fsa 96.94 1309 16209 2284 "R,154" Lib5_Plate173_A01.fsa 99.85 367 2204 2304 "R,155" Lib5_Plate173_A01.fsa 101.21 98 361 2314Notice that when you do fingerprinting your sample file name should include a library name, a plate number, and a well position. It is essential that you are consistent with sample naming, in particular, for each project you want to analyze, fingerprints library, plate number, and well position should appear in a fixed position. I.e., in the sample above the library is "Lib5", plate is "Plate173", and well is "A01". Keep in mind that the output of FPB is the input of FPC in a subsequent assembly and that FPC allows only short names for fingerprints (see FPC manual): therefore in the example above it could be wise to tell FPB that the library is specified at character 4 ("5"), the plate number is specified at positions 11-13 ("173"), and that the well is specified starting at position 15 ("A01").
FPB takes in input all Genemapper exported files with a certain extension that you must specify in input. The file extension can be, for example, .dat or .txt. Files must be located in the same directory from where FPB is run. In the same location must also be present a vector file and a parameters file.
The vector file is used to screen vector bands. If you do not want to screen vector bands then leave the vector file empty. The vector file should be in the following format:
#Vector File Red 90.82 102.77 294.96 -1 Green 177.51 -1In the above example there are 3 red bands and one green band. If you have yellow and blue vector bands you should specify them as well. Remember that different dyes must be divided by the terminating character -1 (this character is used for compatibility format with FPC size files).
FPB parameters
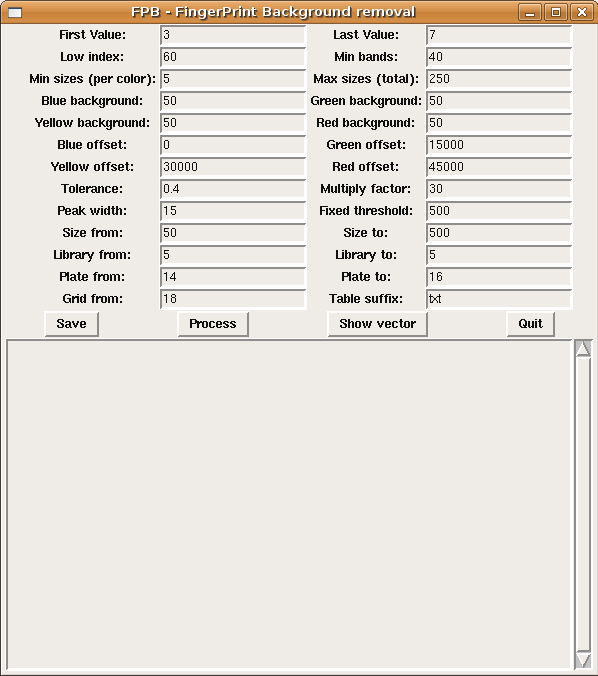
"First value" and "Last value" are the first and last sorted peaks to be used for true peaks determination (see the paper for details). If no offscale peaks (i.e. dye-blobs) are present then "First value" should be 1, as the highest peak is already usable.
"Low index" is the first position (referred to as sorted peak heights) where a true peak should not be found.
"Min bands" is the minimum number of true bands in a clone. "Min sizes (per color)" is the minimum number of true bands per each dye. If any of the dyes does not contain at least this minimum number of true bands then the clone is discarded.
"Max sizes (total)" is the maximum number of bands that a clone can contain, otherwise it is discarded and considered contaminated (too many bands).
"Blue/Green/Yellow/Red background" is the minimum height for peaks to be possibly true, below this threshold they are always considered as background.
"Blue/Green/Yellow/Red offset" is an offset used to map data to .sizes file format.
"Tolerance" is the maximum tolerance of error for a band when compared to vector bands.
"Multiply factor" is used to map data to .sizes file format, each band value is multiplied by this value and then the corresponding offset is added.
"Peak width" is used to remove wide area peaks. In particular, those peaks with ratio area/height greater then this value are discarded.
"Fixed threshold" is used to set a fixed threshold for particular clones with few true peaks. In our experience we got ribosomal clones with a particular patter: few bands and very high signal; computing a correct threshold when few true peaks are present is more difficult, therefore a fixed threshold is used in such cases.
"Size from" and "Size to" is the range of band sizes to be considered.
"Library from", "Library to", "Plate from", "Plate to", and "Grid from" are used to detect library, plate number, and well positions in the file name.
"Table suffix" is the file suffix of text table files to be analyzed: all files with this extension in the directory from which FPB is run will be analyzed, therefore you should choose carefully a file extension for table files.
All these parameters are saved in a text file, parameters.cfg.